BrainOmics 2.0
REGISTRATION CLOSED
BrainOmics – computational approaches to single-cell multi-omics in neuroscience
Category: Course
Location: Human Technopole, Milan, Italy
Date: 18/11/2024 – 22/11/2024
Fee: 700 € (Academic), 1500 € (Industry)
Target Audience: The course is open to up to 20 scientists. Participants are expected to be mostly bioinformaticians and computational biologists with (at least) a basic knowledge of omics techniques. The fee includes all costs related to the on-site activities (including a workstation for each participant), and meals. Accommodation is not included.
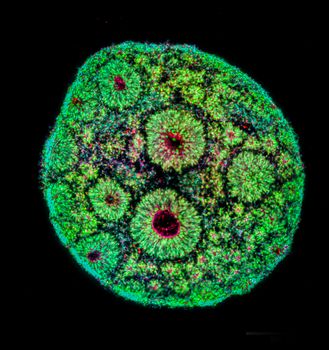
Image: Human cortical brain organoids at 50 days of differentiation displaying the typical rosette-like structures characteristic of the developing human brain. Credits: Aurelio Ortale, Oliviero Leonardi (Testa Group, HT).
Course Overview
BrainOmics 2.0 represents a unique interdisciplinary hands-on course on single-cell multi-omics computational analysis with a focus on the brain and its disorders and builds on the experience of the 2022 edition, with the addition of integrative sessions on functional genomics.
Single-cell omics technologies are breaking new ground in neurobiology by substantially increasing the precision and resolution with which the complex cell populations of the nervous system can be characterised. Approaches that profile several layers of information (genome, epigenome, transcriptome, proteome, spatial location) allow to generate data of unprecedented depth on the molecular state of the diversity of cells composing the nervous system.
This increase in data volume and complexity generates as many opportunities as new analytical challenges.
This compact course aims at empowering participants in mastering key computational tools for the analysis of single-cell omics datasets, starting from individual molecular layers to then tackle their integration, alongside providing a theoretical overview of the impact of these technologies at the leading edge of neurobiology.
Target Audience
This course is aimed at bioinformaticians and computational biologists with expertise or interest in neurobiology and (at least) a basic knowledge of omics techniques.
The core of the course will be centred around hands-on data analysis sessions. A basic understanding of Unix command line, Python and/or R is required. Previous experience with single-cell analytical workflows will be considered as an added value to harness the most from the course training sessions.
SPEAKERS & INSTRUCTORS


Contact
Call for Sponsorship No.05 - BrainOmics course 2024: computational approaches to single-cell multi-omics in neuroscience






















